Rotea Protocol Review Tool - Context Help
Purpose:
Load a Rotea protocol and compare it against best-practice guidelines.
Intent:
Assist Rotea users in building robust, reliable applications.
What It's Not:
This tool does not create or modify protocol files. To make changes, use the Rotea Protocol Builder.
Overview
This application:
Parses the Rotea protocol file created by the Rotea Protocol Builder.
Analyses the protocol through four simulation functions:
- structure of the raw protocol definition
- Volume modelling through algorithms
- Detailed volume simulation emulating the sequence
- The interaction of cell like particles to the chamber conditions.
Creates a report recommending changes for improvement of the protocol based on guidelines.
- Guidelines are a suite of 'best practice' behaviours developed through a risk-based approach.
- As the simulation is progressed, conditions where a guideline can be assessed are traversed.
- Hard coded checks compare the conditions to the guideline settings.
- A guideline report is generated if non-compliance is recognised.
Supports the user to create a documented change plan
- Enables iterative development
What to Expect
It is common to find guideline issues in protocols.

If the volume simulation is completed with no guideline conflicts,
our rather concerned face becomes:

The goal of this tool is to help you achieve this happy face - In the protocol and on the face of your QA Director.
Beyond the Basics: Exploring Insights
This application isn’t just about compliance; it also provides insights into protocol performance:
Parameters: Exploring settings to align with process requirements.
Reagent Requirements: Estimate the reagent volumes and output bag capacities needed.
Priming Functions: Evaluate priming performance and identify remaining process hazards.
Cell Input Variation: Analyze how variations in input bag volume and cell concentration affect performance.
Cell Dynamics: Visualize the accumulation and elutriation of cells within the chamber.
Cell Loss Patterns: Identify sources of systematic cell loss.
Cell Recovery Score: Evaluate the process success score that quantifies cell recovery performance.
The Role of Guideline Recommendations
The primary purpose of these activities is to evaluate the protocol against best-practice guidelines.
- As the simulation progresses, the tool identifies steps where protocol behavior deviates from guidelines.
- Guideline Reports are generated for each detected deviation.
- These reports are accessible alongside the step listing, allowing you to review and act on the findings.
Interacting with Guideline Reports
When you open a Guideline Report, you can:
- Add Comments: Document observations or decisions.
- Suppress the Guideline: Acknowledge the deviation for the current step if it is acceptable.
- Commit a Recommendation: Accept the guideline’s suggestion as a planned protocol change.
Committed recommendations appear in the Change Plan Report, which:
- Aggregates all planned changes for easy tracking.
- Can be printed to PDF for documentation and quality assurance purposes.
Iterative Refinement
This guideline interaction process enables continuous protocol improvement:
- Run the simulation and review the guideline reports.
- Commit or suppress recommendations based on process requirements.
- Edit the protocol using the Rotea Protocol Builder.
- Re-analyze the updated protocol to verify improvements.
Over time, this iterative approach ensures the protocol either:
- Meets best-practice standards, or
- Includes justified deviations that are documented and approved.
The ultimate goal: A robust, well-documented protocol that meets both operational needs and quality standards.
Getting Started - Your first pass through the workflow
This guide will walk you through the basic workflow to help you navigate your first protocol analysis. We recommend starting with a standard test protocol to get familiar with the process and outcomes. However, you can also use your own protocol if you prefer.
Workflow Overview
The analysis follows these key steps:
- Load the protocol and run an initial structure check.
- Review the protocol definition to prepare for the volume simulation.
- Run the volume simulation to model fluid behavior.
- Prepare for the cell simulation by verifying necessary parameters.
- Run the cell simulation to analyze cell behavior.
Step 1: Load the Protocol
- Click the Protocol Button.
- Select your protocol file. For detail: refer to Protocol Loading Errors.
- If successful, the next step will become available.
- If an error occurs, refer to Protocol Loading.
Step 2: Review Modify File Data
(This step is skipped by the application if no previous data has been saved.)
- Click the Modify Button.
- Review the modification settings.
- Adjust preferences or use Yes/No to All to continue.
- For more details, see Mod File Data Review.
Step 3: Initialize Volume Simulation
- Click the Volume Init Button.
- Adjust register settings or bag volumes if needed.
- Continue until you see this indicator:
- If adjustments don’t resolve the issue, see Volume Simulation Initiation.
Step 4: Run the Volume Simulation
- Press the Volume Sim Button to run the simulation.
- The system will display progress and any detected guideline issues.
To Review Results:
- Select Kit View to see fluid and air distribution in the system.
- Select List View to inspect bag fill volumes.
Reviewing Simulation Outcomes
- Guideline issues appear alongside the step list.
- Click on any step to review associated reports.
- Open the Information Tool for a summary report, which can be saved as a PDF.
- If the simulation fails, refer to Volume Simulation Troubleshooting.
Step 5: Initialize Cell Simulation
- Click the Cell Init Button.
- The system will review available data for the cell simulation.
- If adjustments are needed, see Cell Simulation Initiation.
Step 6: Run the Cell Simulation
Press the Cell Sim Button to start the simulation.
The button will change to:

Progress indicators and the kit schematic will update as the simulation runs.
Note: The simulation time varies depending on the number of cell types.
Step 7: Explore the Results
Analyze Cell Distribution
Open the Information Tool:
View the Cell Reconciliation Report to track cell distribution.Inspect Final Cell Positions:
- Go to the last step in the process.
- Open Kit View.
- Select each bag to see the composition of cells and media.
Visualize Fluid Dynamics
Run the Animation:
Select the Run Arrow to visualize fluid movement through the kit.Pause or Stop Animation:
Press Stop to halt the animation at any point. (There is no pause feature.)
Chamber Insights
- Open the Chamber View to examine:
- Cell Accumulation: Track how cells collect and separate in the chamber.
- Chamber Capacity: View a chart displaying the cell-retention potential under current conditions.
Step 8: Review and Adjust Parameters
Return to the Volume Init Screen to review suggestions for:
- Bag starting volumes.
- Parameter settings.
Click on any value to adjust it.
Reminder:
Any change here will automatically reset the Volume
and Cell simulations.
Next Steps
With your initial simulation complete, you can now:
- Investigate guideline reports and apply suggested mitigations.
- Generate PDF reports for documentation.
- Modify the protocol in the Rotea Protocol Builder.
- Re-run the analysis to confirm improvements.
Protocol loading
- The protocol file is never altered by the application.
- It is important your protocol file is located to a working area in your computer file system.
- The simulation will create a subdirectory at the protocol location to save the run log and report files.
- Select the 'Protocol' button at any time
The first time a protocol is loaded the 'Modify' is skipped since no changes have been saved
Once the protocol is loaded you can review, the operations in Kit view and any parameter settings in List View. Select a line in either view to observe the trigger and any repeat functions.
Protocol description information
Press the 'information' tool
![]() when the protocol is loaded to review the description data from the
protocol.
when the protocol is loaded to review the description data from the
protocol.
- To avoid capture of potentially sensitive information, this information is only available when the protocol is first loaded.
- There is no facility to print it.
- To re-display this information at any time, press the Protocol button, and select 'No' to opening a new protocol.
Protocol Loading Errors
- Access privileges for the location of the protocol file need to be resolved, or a copy of the protocol file must be placed in a working directory
Selected File cannot be analysed
- There is likely some variation in the json file details of this file, potentially from an early release of the Rotea protocol builder.
- Verify you can open it with the Rotea Protocol Builder software and re-save it from that application. Then re-try opening it.
- If you cannot edit the file with the Rotea Protocol Builder, the application cannot interpret the file.
Protocol review errors:
![]()
- There are errors in the protocol recognised by the preliminary analysis.
- Normally a description of the error will be displayed.
- A common fault, for example, is failure to include a pause step for the final step.
- The information box will display a line highlighted with the error summary.
- The Guideline Reports tool is illuminated also.
- You can select the Guideline reports tool or double-click on the information box line to display the guideline commentary in the guideline report.
- If an error is raised, the guidelines are strongly recommending this issue should be corrected
- It is recommended you 'commit' the guideline as a change activity.
- So you can proceed with the analysis, you can 'suppress' the guideline for this step.
- If you disagree with this guideline assessment, you can disable it, or change the alarm level, in the Guideline editor.
Modify File
This file holds settings you have used to analyse the protocol.
- It is created when you first make any changes.
- It is located in the working file directory.
- When you open a protocol, the application opens the mod file if it exists.
- The 'Modify' control is enabled if the file is found.
- Otherwise, default settings are drawn from the protocol and the 'Volume Init' control is enabled.
Protocol Changes
The working directory and modify file are linked to the filename of the protocol.
Any change to the protocol filename will create a new working directory independent of the previous protocol.
When a protocol is saved by the Rotea Protocol Builder, a timestamp is included.
If this application sees the timestamp has changed when opening the protocol, the guideline settings in the modify file are cleared.
Modify screen
- Changes or additions to the protocol derived settings are saved in the modify file and displayed here.
- You can choose between the new settings or return to the protocol derived data.
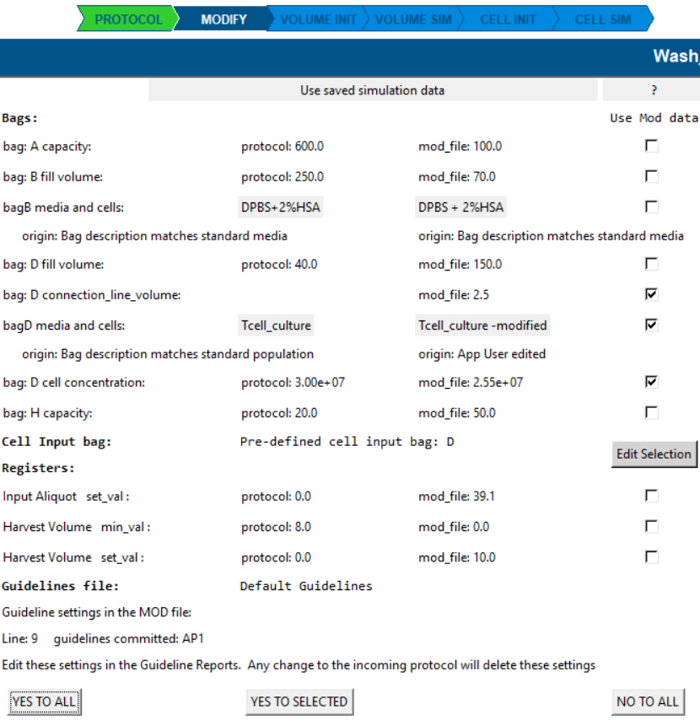
- You can return to this modify screen and change preferences at any time.
- Changes will clear the simulation.
- Click on the population labels to view the population details.
Cell Input Bag
- This application needs to know which bag contains the starting population of cells/particles.
- If it recognises a cell population in the protocol data it will use that bag.
- Otherwise, you will be asked to choose which bag.

Why do we need to know the cell input bag?
- Knowing which bag contains the cells lets the application understand where priming stops and processing begins.
- Once processing begins, drawing from, or pushing into untested fluid lines raises a guideline warning.
- Priming operations before processing can be reviewed in isolation for completeness.
- Once volume simulation is completed, a summary of the priming operations is presented in the information tool.
- In some processes other bags may contain particles such as magnetic beads. The primary cell product bag is the focus here.
Working file directory
- A local file directory is created when a protocol is loaded to receive and coordinate these documents.
- The name of this directory is highlighted in the information tool.
- It comprises the first 6 letters of the protocol name plus a hexadecimal checksum of the entire name.
- If you do not re-name your protocol through editing, the analysis outputs will return to the same sub folder.
Initialise Volume Simulation
The volume simulation requires more information
than the minimum data in the protocol file.
This information includes:
- Starting fluid volumes: The initial fluid volumes for each bag, along with their capacity.
- Parameter settings: Parameters that influence fluid transfers throughout the process.
The Initialise Volume Simulation function checks
that these parameters are defined sufficiently.
If valid data is detected, the Volume Simulation
can proceed.
Note: The simulation can still run with suboptimal or incorrect settings — this is intentional.
Observing poor settings in action can provide valuable insight into protocol behavior.
Bag volumes and parameter settings can be adjusted directly by selecting the relevant data field.
You can also edit bag volumes by selecting the corresponding bag in Kit View.
Any changes made to these settings will reset any existing volume simulation results.
Protocol Parameters
The Rotea Protocol can use parameters to control:
- Step timing
- Fluid volumes
- Loop iteration counts
Run time settings for these parameters are not stored the
protocol file.
The protocol file does carry minimum and maximum
limits for the run-time setting.
How to Edit Parameters:
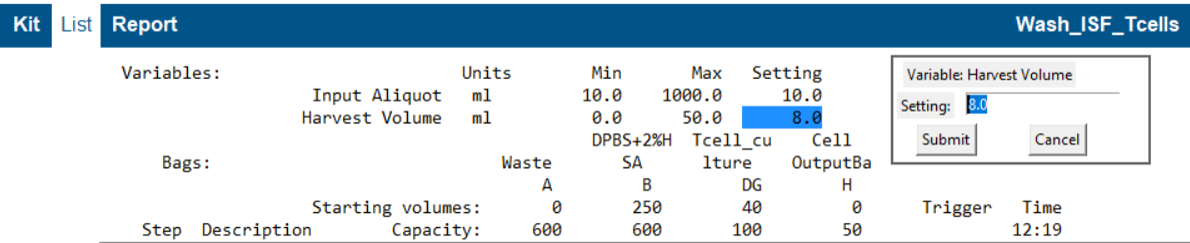
- Open the List View after loading a protocol.
- Click on any parameter, minimum, or maximum value to edit it.
- Parameters cannot exceed the defined limits.
- Adjust the max/min values first if you need to set a parameter outside the current range.
Changing a setting will clear the existing volume simulation, simply re-run the simulation when ready.
Parameter applications
Setting the parameters for a program can be non-intuitive with the potential to impact process reliability if poorly selected at run time.
Good practice is to define maximum and minimum limits in the protocol for useer guidance.
TBD suggest maximum / minimum limits based on this analysis
The application tries to comprehend how each parameter is being used to check guidelines for that activity. Three common applications:
1. Final Product Volume (Harvest/Recovery Step)
Many users take advantage of Rotea's ability to deliver the final
product in a small volume.
It is common to have a parameter to control the harvest
volume.
For example if the setting is say 0.3ml:
- Rotea will deliver that 0.3ml past the output valve if the step is a 'Harvest Step'.
- Before that volume gets to the output vessel:
- it must be pushed through the tube in the kit from the valve
- and the tube to output bag or vial.
- If these volumes are not budgeted, then no final product will get to the bag.
Guideline AV19: Warning Is triggered when no fluid reaches the output bag in a recovery step.
2. Cell Load Volume (Multi-Bite Sequences)
Protocols often use multi-bite loops to load
cells gradually, preventing the chamber from being
overwhelmed.
A typical multi-bite loop involves:
- Loading a volume of cell product -- controlled by a parameter setting.
- Washing the captured cells.
- Recovering the cells into an output or intermediate bag.
Loop behavior depends on:
- Input bag volume:
- The loop repeats until the input bag bubble sensor detects the input bag is empty.
- Bite size settings:
- Determined by data parameter setting.
- These factors affect
- how many loop cycles occur
- the volume of the last bite.
Why This Matters:
A low volume last bite results in small numbers of cells in the chamber.
- This disrupts bed formation, elutriation and washing steps.
- Leading to cell loss, lower final concentration, and poorer separation.
A low bite occurs because an extra loop cycle occurs before the bubble sensor trigger.
- One extra loop cycle will use more reagent, require more waste and output bag capacity
- The final product is diluted unnecessarily by one more harvest volume (with no cells in it).
Finding the right parameter setting
The parameter setting must respond to the input bag volume to keep the last bite volume within a target range.
For example:
- If the target volume per bite is 100 mL,
- The final bite should be at least 80% of that target
- (i.e., 80 mL or more - based on the guideline AV8 threshold setting).
And further, the final bite volume should be less than say 95% to avoid the risk of another bite.
So we want a parameter setting that places the final bite size between a target minimum and a maximum.
Guideline AV8 monitors final bite volume.
It issues a warning if the simulated last bite size is below the guideline threshold.
3. Dilution Volume (High Cell Concentrations)
Processes involving whole blood or leukopak material often need a dilution step to:
- Reduce the incoming cell concentration so the chamber function is not overwhelmed.
- Adjust the media density to support selective elutriation.
The cell simulation is a good way to explore these issues. By way of explanation at the time of writing; Cells flowing into the chamber take up a proportion of the fluidised bed capacity to retain cells in a region of the chamber. If the incoming cell concentration is very high, there is no capacity of the local fluidising environment to retain any cells. This may diminish further into the chamber, so if the number of cells is small enough, the bed can settle down once the inflow of cells stops. So a small bite volume is one approach. If using this approach, be aware that the bubble trap contains around 5ml of incoming material that needs to run into the chamber before a wash fluid enters the chamber.
- The cell simulation can detect when the number of cells in the chamber exceed the chamber capacity.
- Dilution may address this issue by reducing the cell count per bite where the bite volume is fixed.
Guidelines monitoring cell operations:
- CL1: Monitors cell concentration entering the chamber.
- CL2: Tracks total cell count in the chamber (available in Volume Simulation if a starting population is defined).
- CL3: Active during Cell Simulation — triggers a warning if cells wash out due to capacity overflow.
Special Case:
- If a dilution step doubles as a priming step and the dilution volume register is set to 0, the input line may remain unprimed.
- This can cause air ingress and process failure.
Guideline AV26 detects and warns of unprimed input lines.
Predictive Tools
The Initialise Volume screen provides several predictive tools to assist with protocol optimization.
1. Predicting Reagent Volumes and Bag Capacity
The tool analyzes the protocol to:
- Predict reagent consumption based on current settings.
- Estimate bag capacity requirements.
- Highlight insufficient volumes for protocol completion.

- Highlighted settings do not meet the predicted requirements.
- Adjust the bag starting volume or capacity to resolve any highlighted issues.
Note: The simulation can still proceed with insufficient volumes, allowing you to observe potential process failures and better understand system behavior.
Capturing parameter setting hazards
Since the protocol does not include run time settings for parameters, the settings on first review can be = 0. (On first review the application will set the parameters to the minimum set point.) An infinite loop can occur in some cases and these raise an error that prevents the simulation from starting. Where a harvest or recovery step is recognised, minimum values are recommended. The warnings and comments are displayed beside the parameter entry and in the information box.
Supporting multi-bite parameter settings
The protocol analysis is used to predict the volume of the last bite of a multi-bite sequence based on the bag volumes and existing parameter settings.
Guideline AV8 compares the estimated last bite volume as a % of the target volume for the step. The protocol analysis seeks to keep the last bite volume between the minimum 'Threshold' and 'Maximum' % settings of the guideline.
If greater than maximum percent there is a risk errors in bag volume measurement might result in an extra unwanted loop.
If less than threshold elutriation and/or washing steps may be compromised.
Setting the parameter value
The protocol analysis displays:
- its expected outcome for the current parameter setting as a number of loops / percent of target volume in the last bite.
- If the percent of target for the current parameter setting is not within the guideline % range, it is highlighted.
- A series of three recommended settings are presented: number of loops / parameter setting.
- The centre of the series reflects the current parameter setting.
- Change the parameter setting to assess different numbers of loops.
- Use one of the recommended volume settings to observe the predicted compliance with the guideline.
- (Check back in with the volume simulation to observe the simulated versus predicted percent outcome at the last cell loading step)
2. Detecting Parameter Hazards
- Protocol files do not include preset values for the parameter settings.
- If no minimum value is set, the parameter setting will default to 0.
Risks:
- Zeroed parameters can cause infinite loops in some cases.
- Harvest/recovery steps require minimum settings to avoid step failure.
The application:
- Blocks simulation start if a potential infinite loop is detected.
- When the protocol is first opened, sets the parameters to minimum setting value, (which can be 0).
- Displays warnings and suggests minimum settings.
The example below illustrates parameters controlling a bite volume and harvest volume.
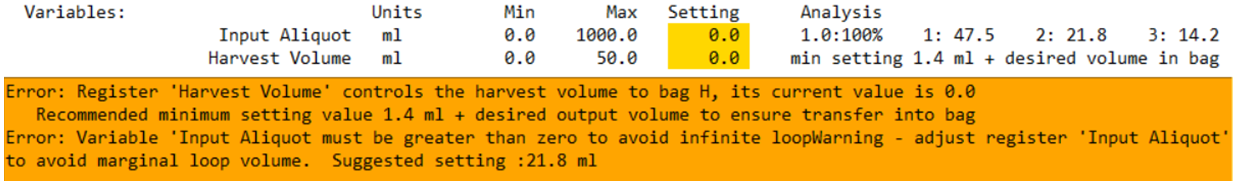
3. Multi-Bite Parameter Setting
The multi-Bite parameter setting tool estimates the last bite volume in multi-bite loading steps. While all elements of the protocol can influence the result, it is specifically directed by:
- The input bag volume.
- parameter settings.
The tool suggests parameter settings that comply with guideline AV8.
Guideline AV8 Monitoring
Guideline AV8 raises a warning if:
- The last bite volume is too small (commonly below 80% of the target volume)
- The final bite is too large, which risks triggering an extra, unintended loop. (The upper limit is typically 95% - AV8 Maximum setting - AV8 can be edited in the Guidelines Editor)
The target volume is the step volume driven by the parameter setting.
Example Display

In this example:
- The parameter setting is 30.0 ml.
- The highlighted field shows:
- The predicted number of loops: 3
- The final bite size: 67% of the target volume. (67% x 30ml = 20.1 mL)
This value is highlighted because it does not comply with the AV8 guideline settings.
- The final bite volume is too small, falling below the 80% threshold.
Suggested Parameter Settings
To the right of the highlighted field, you'll see three suggestions for parameter settings.
Each entry shows:
- The Expected Loop Count.
- The corresponding parameter setting.
In the example above:
- 3 loops : 28.2 ml suggested register setting
- 4 loops : 20.9 ml
Adjusting the parameter Setting
If we set the parameter value to 28.2 ml, the display refreshes:

What Changed?
- The highlight disappeared indicating compliance with AV8 guidelines.
- The expected loop cont is now 3.
- The final bite volume is estimated to be 85% of the target volume. (28.2 ml = 85% x 23.9)
Important
- This estimate relies on how well you know the input bag volume .
Understanding the Display Dynamics
When you adjust the parameter setting, the display dynamically updates to reflect the predicted loop behavior.
Display layout
- The current predicted loop count is shown in the center.
- The suggested parameter settings for one loop less and one loop more are displayed on the left and right, respectively.
How to Adjust the Setting:
- Modify the parameter setting by clicking on the value and servicing the displayed edit box.
- Observe the predicted loop count - The suggested volume for this loop count is in the center.
- Apply the suggested parameter setting once the desired loop count is displayed.
Choosing the Right Loop Count
In many cases, fewer loops are preferred because they:
- reduce sensitivity to input bag volume measurement variability.
- simplify the overall process, reducing overall operational risk.
Understanding the Role of Looping
looping is primarily used to manage:
- Large input volumes of material.
- High cell densities that need to be processed in smaller increments.
Risks of Inappropriate Loop Parameter Settings
- Target Volume Too High:
- There may be more cells than the chamber can hold for each bite, resulting in losses to waste.
- Elutriation steps cannot perform well because there are too many cells to allow the bed to expand without overflow.
- Target Volume Too Low:
- There may not be enough cells to form a robust bed with low concentration input material.
- All subsequent steps that rely on the presence of a bed, will be disrupted.
The cell simulation monitors cell overflow issues. For more information, refer to cell concentration ranging
Bag Fill Range
The register controlled bite volume settings assume a known input bag volume.
At run time however, accurately measuring the input bag volume can be notoriously difficult.
The 'Fill volume range' displays the bag volume matching the upper and lower thresholds.
- The format is:
final bite volume % / input bag starting volume
- The format is:
If the actual bag volume is within these bounds, the process will meet the guideline AV8 recommendations.
These values represent a volume measurement tolerance.
- These values are only displayed when the parameter setting predicts a final bite volume within guidelines.
Input Bag Volume Ranging
Changes to the input bag volume need to be accommodated as a normal part of run-time operations.
The input volume slider tool helps you to explore the effects of changing start volumes:
- Allowing you to adjust the input bag volume.
- Dynamically updating predicted reagent use and the impact on recommended parameter settings.
How to use the slider controls

Slider control buttons
These buttons are enabled whenever you move a slider.
Reset Changes:
- Restores the slider to its original position.
- Discards any unsaved adjustments.
Save Changed Settings:
- Saves the new settings to the Modify file.
- Changes are then retained for future simulations
Tip: Click on the maximum or minimum slider values to adjust the allowed volume range.
These values are saved in the Modify file.
Once exploration is complete, set the ranges to the bounds expected for run_time operations.
Cell Concentration Ranging
The total cell concentration of the input bag can be adjusted with a dedicated slider.
When you move the slider:
All cell concentrations in the input bag are adjusted proportionally.
Real-time updating predicts the impact of the new setting.
Any change of the slider resets the volume and cell simulations.
After adjusting the slider, rerun the volume and cell simulations to observe the updated results.
The slider button functions are identical to the volume slider
How the Tool Works with Guidelines
Changing cell concentration affects two guidelines that can be assessed at this time:
Guideline CL1
- Monitors the cell concentration of product entering the chamber.
- If a parameter is available to control
dilution, the tool will:
- suggest a setting to keep the concentration within the guideline threshold limits.
- If no register is available:
- A warning is raised, alerting you to a
potential chamber loading failure.
- This occurs when cells are washed through without dispersing properly into the chamber.
- A warning is raised, alerting you to a
potential chamber loading failure.
Guideline CL2
- Monitors the total number of cells being loaded into the chamber by a step.
- Compares the predicted cell count to the guideline threshold setting.
- If a bite volume register controls the step:
- The tool may suggest lowering the bite volume if the predicted cell count exceeds the threshold..
- The number of loops suggested will increase accordingly.
- If no register is available:
- A warning is raised to highlight potential chamber overloading
- where cells are washed through to waste.
- A warning is raised to highlight potential chamber overloading
Methodology
- Adjust the cell concentration slider's maximum and minimum limits to real-world estimates for your process.
- Use the slider to simulate real-world variations in cell concentration.
- After adjusting the slider, rerun the volume and cell
simulations to observe the updated results.
- In particular look for simulated chamber overflow, guideline CL4, arising in the cell simulation.
- Consider the need for:
- A minimum value for any dilution step parameter setting.
- An additional dilution step or increased priming volume.
- A maximum value for the bite volume register setting.
Volume Simulation
The volume simulator simulates fluid movement, identifies bubble sensor staus, and determines the triggers that control the end of each step.
It emulates loop controls to generate a sequence of steps based on the defined protocol.
The volume simulator estimates:
- The fluid volume transferred during each step.
- The time duration required to complete each step.
The List View displays:
- The fluid volumes in each bag.
- The trigger type and duration for each step.
Selecting a step reveals detailed information about its triggers and loop controls in the information box.
The volume simulation's primary goal is to mimic process conditions for guideline analysis.
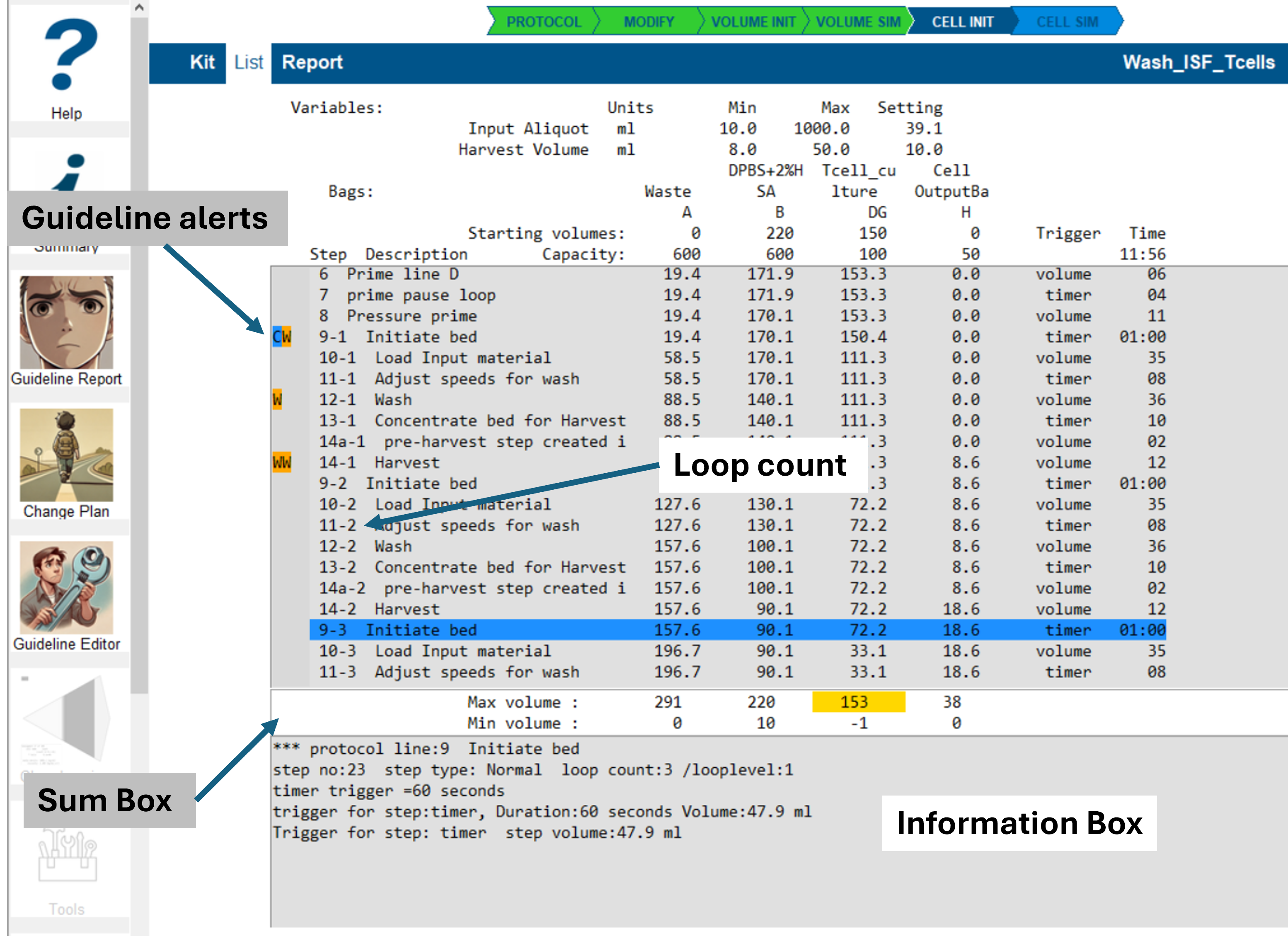
Guideline Deviation Display
Guideline alerts are highlighted as color-coded text symbols to the left of the step list:
- A - Advice
- W - Warning
- E - Error
- C - Committed for change
To review these reports:
- Switch to Kit View and select any step with highlighted symbols.
- The information box will display a color-coded summary of the associated guideline reports.
- Double-click any line to open a detailed view of the corresponding guideline report.
Tip: For more information, see the Guideline Reports section.
Volume Simulation Results
The image above shows a typical volume simulation result for a protocol with a single active loop.
The blue-highlighted line indicates the selected step, which was chosen by clicking on it.
Information Displayed in Each Line
- Guideline Alerts: Displayed if any guideline issues are detected.
- Protocol Line Number and Loop Count: Displays
the protocol line number and loop
count.
(Nested loops may display multiple loop counts.) - Step Description: A brief summary of the step's operation.
- Bag Volumes: The fluid volume in each bag at the end of the step.
- Trigger Type: The trigger type
that determined the step's volume.
- This information is displayed in the information box and can also be inferred from step-to-step volume changes.
- Step Duration: The step's duration in HH:MM:SS format (Hours:Minutes:Seconds).
Information Box Details
The information box displays additional step-specific information, which is also stored in the log file in the working directory.
Displayed Information Includes:
- Protocol Line Number and Description:
- Identifies the protocol line being executed.
- Step Number, Step Type, and Loop Level
Tracking:
- Protocol lines may be repeated multiple times within loops.
- Step numbers indicate the execution sequence within the protocol.
- The step type is defined in the protocol configuration.
- Harvest Step Adjustment:
- For harvest steps, the simulation inserts a pre-harvest step to simplify process logic.
- Step Triggers:
- Multiple triggers can be associated with a step.
- The Trigger for Step field shows the trigger type and final trigger volume for the step.
Understanding Negative Volumes
When fluid is drawn from a bag, the Rotea system determines when the bag is empty by monitoring a bubble sensor.
Bubble sensors are positioned at the end of the fluid line connecting the bag to the kit.
As a result, the bag volume may be drawn down below zero representing the fluid in the connecting line.
- In some cases, it may be necessary to retrieve the remaining fluid from the kit to the bubble sensor.
- To achieve this, a controlled 'draw-down' step may be used to move fluid past the bubble sensor.
- As a result, the bag volume becomes
even more negative.
- This action will trigger a guideline error; however, since it is intentional, the guideline can be disabled for this step.
The volume simulation intentionally allows these negative values to emerge and propagate through the simulation.
Guidelines continuously monitor the process and raise warnings or errors if potential issues are detected.
Troubleshooting Failed Volume Simulations
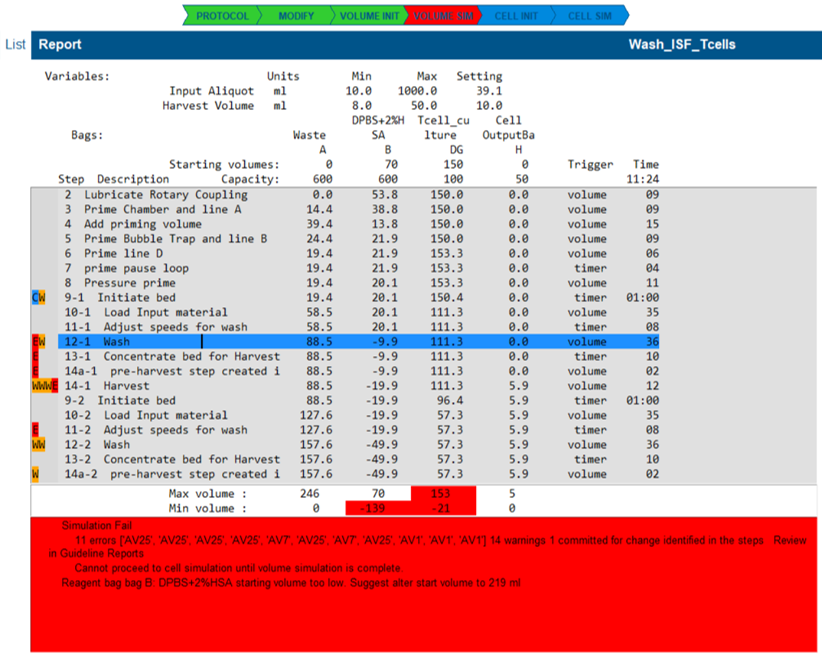
A volume simulation will fail if one or more guidelines raise an error condition.
- the illustrated state highlights many guideline errors that have occurred because the reagent ran out.
Check for Reagent Shortages
- Open the List View and check the summary box for a reagent shortage message.
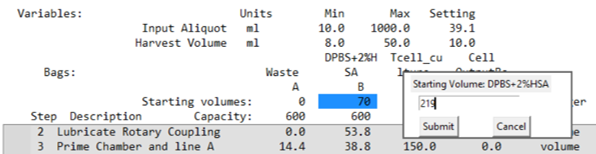
- The example report indicates Bag B is too low and suggests a new starting volume.
- If a shortage is detected:
- Adjust the bag start volume by clicking the value setting and entering a new amount.
- Re-run the simulation by clicking the Volume Sim button.
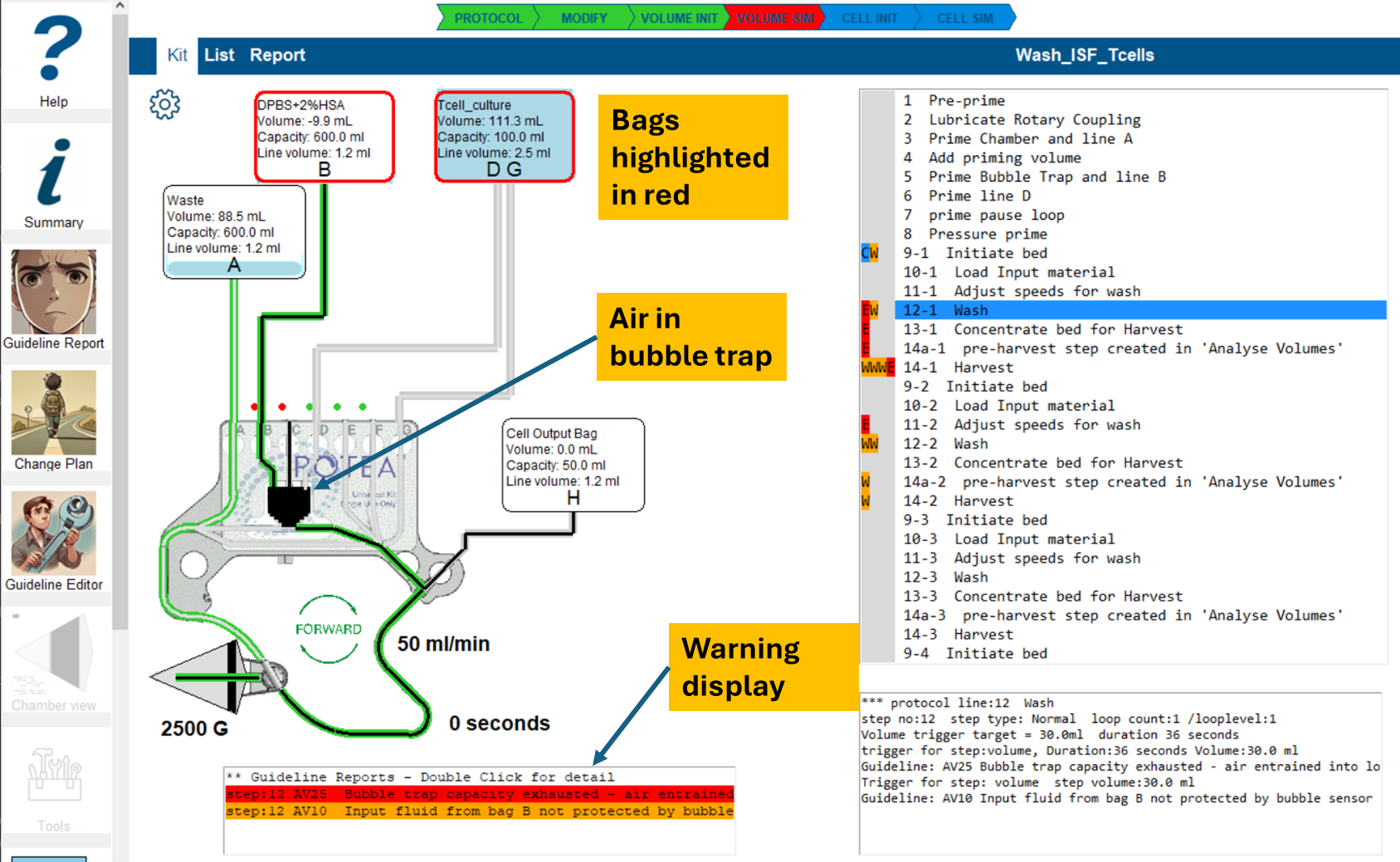
Review Guideline Alerts
- Look to the left of the step listing for colored guideline indicators.
- Identify the first step with a red error indicator.
- Switch to Kit View. (Refer to image below)
- The warning display, displays a summary line for each guideline alert of the selected line.
To investigate further:
- Double-click the guideline summary line to open the [detailed guideline report](#Reviewing guideline conflicts in your protocol).
To resolve the issue:
- Select 'Commit' to add the recommendation to the protocol change plan.
- Select 'Suppress' to disable the guideline temporarily.
- Save the changes to apply the selected action.
- Return to the Volume Init screen.
- Re-run the volume simulation to verify if the issue has been resolved.
Kit view in volume simulation error state
This view also highlights visualization of the air in the kit, (shown in black,) at the end of the step.
Cell Simulation Initiation - Cell Init
The protocol settings and bag contents are reviewed to check if there is sufficient information to proceed with the cell simulation.
- All bags with a starting volume >0 must have media defined.
- For a cell simulation, the 'cell input bag' must have a
particle/cell population defined.
- Other bags can have particles present also.
- Missing information is highlighted in Kit view with the offending bags in red.
- The bag population data can be edited from this point.
Bag contents can be edited in Kit view any time after the protocol has been loaded.
All changes to bag contents are saved in the Mod file for re-use.
- If you edit the bag contents, re-try the 'Cell Init' function.
- A message will advise when the cell simulation can proceed.

Editing bag volume data
- In Kit view select the bag to be edited.
- The top window displays the label, starting volume, bag capacity and line volume for direct editing.
- Any change to the volume settings will reset the volume simulation.
Editing bag population data
- In Kit view select the bag to be edited.
- If the bag starting volume >0:
- Below the label is the description of the bag population (cells and media)
- Edit button displayed
- Select the edit button to display the existing population data for the bag.

Using standard populations
- A description of the population is displayed at the top with a
'Select Population' button
This allows the entire population to be created from a standard group.
The bag population selected this way can be edited in detail, independent of the standard group.
Pick on the drop-down box to display and select from the available standard groups.
The bag population is now displayed with the standard group details.
Press 'Submit' to confirm the editing process and return to the kit view.
It is not required that a standard population be selected. Reagent bags for example do not require a particle population.
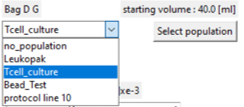
Note the 'protocol line 10' entry in the list. This refers to a population found in the protocol builder process simulator for line 10.
Defining or editing the media in the bag
- When the bag population data is displayed the 'Media' section displays a summary of the existing bulk density and viscosity.
- The box below displays any media description and the percentage contribution to the total volume.
- An 'Add media' line is also displayed.

Edit media item
- Select the media item from the list in the display box
- A window allowing you to edit details of the media is displayed.
- If more than one media is in the population a 'Delete' button is available to remove this item.
- Change the description and properties as you wish.
- Set the 'Percent' item to your target for the media.
- When the changes are submitted, the percentage of any other media components will be adjusted to honour this value.
- Submit changes returns to the population display window but changes are not locked in until the population window is 'Submitted'
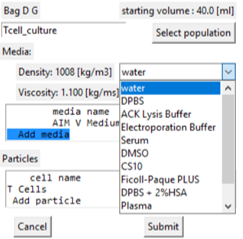
Adding media to the population
- By adding media we are changing the components in the media - not changing the volume.
- Select the 'Add media' line in the display box.
- A drop-down box of the stored standard media types is displayed for selection.
- If selected the media edit window is displayed.
- The Percent of this media in the final volume needs to be entered.
- Default value is to be equi-proportional to the other media types.
- The media description and properties can be edited at this time.
- Submit changes returns to the population display window but changes are not locked in until the population window is 'Submitted'
Defining or editing the cells / particles in the bag
- When the bag population data is displayed the 'Particles' section displays a summary of the existing particle population.
- For each existing cell type, based on the starting volume for
the bag, (top right of population window,):
- the cell count is displayed based on the cell concentration.
- the percent of the total population count is displayed.
- An 'Add particle' line is also displayed.

Edit cell / particle item
- Select the cell item from the list in the display box
- A window allowing you to edit details of the cell type is displayed.
- A 'Delete' button is available to remove this item from the population.
- Change the description and properties as you wish. Refer to Particle Properties for more detail.
- The 'Concentration' entry is specific to this use of the cell type.
- The Colour button can be used to set a visualisation colour for
the cell type.
- The colour and visibility of each cell type can be set dynamically using the Kit Settings window.
- Submit changes returns to the population display window but changes are not locked in until the population window is 'Submitted'
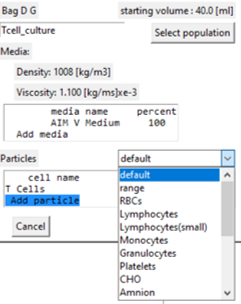
Adding cells / particles to the population
- Select the 'Add particle' line in the display box.
- A drop-down box of the stored cell / particle types is displayed for selection.
- If selected the particle edit window is displayed.
- The immediate focus is to specify the cell concentration that is 0 by default.
- The description and properties can be edited at this time, Refer to Particle Properties for more detail.
- Submit changes returns to the population display window but changes are not locked in until the population window is 'Submitted'
Particle Properties
Concentration : units 'cells/ml' using scientific notation: 1 million cells/ml = 1.e6, 150 million cells/ml = 1.5e8
Cell Density : units 'kg/m3' = 'grams/liter' A Minimum and Maximum value can be specified to reflect normal variation
Cell Diameter : units 'micron' A minimum and maximum diameter can be specified.
Compensation Factor : Ratio = 1.0 +/- 0.2 say to direct the particle behaviour in the model to reflect your experience:
- more than 1 makes the cell relatively heavier more reluctant to elutriate.
- less than 1 makes the cell lighter and more inclined to elutriate.
- Red blood cells seem to behave more realistically with a compensation factor = 0.8 for example.
Select the histogram image to review the particle size distribution.
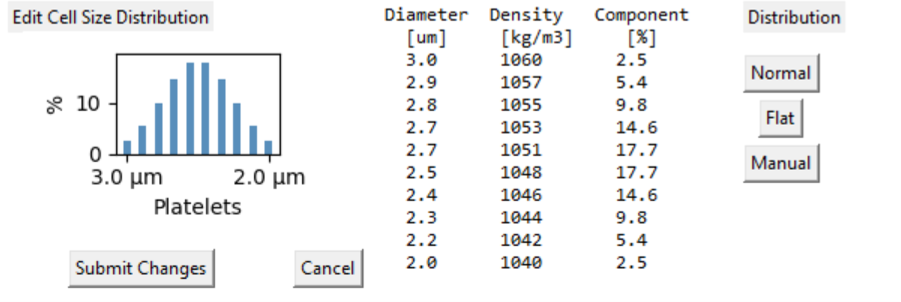
Particle Size distribution
Cells exist as a population of shape, size and density. The
simulation assumes all cells are spherical.
Size and density simulation supports differential cell selection
within a population.
So some cells can accumulate in the chamber while other cells are
elutriated out for example.
Physical measurements suggest most cells are represented by a normal
distribution which is the default.
It is not likely that the largest cells will have the heaviest
density for example. On the other hand definition of a cell type may
include multiple variants of a generic cell so a purely normal
distribution is less appropriate.
Be aware that the distribution is based on (diameter cubed) x
density. This reflects the relative behaviour of these cells in the
chamber and how they interact with other cells in the chamber. The
table displays the diameter and density for with each 'bin' in the
distribution. Select different distribution types as you see
fit.
The 'Flat' distribution is useful to explore the effects of CF ratio
changes on the process outcomes. The 'Manual' function enables
sliders to create a flat topped distribution that may reflect
multiple cell species. The distribution specification for each cell
type is saved in the 'Mod' file.
Cell simulation
The purpose is to explore how the cells move around the kit in response to the process.
Creating the opportunity to recognise unexpected outcomes from the protocol.
Highlighting guidelines that rely on cell population calculation to be assessed.
- For example, how do we know if we are trying to put too many cells in the chamber?
Predicting the protocol performance in terms of yield.
Identifying where we might be losing cells systematically for want of minor protocol changes.
It is important to recognise this simulation of cells in the
bags, kit and chamber is intended to highlight how the protocol
effects the movement of cells rather than any absolute
representative model.
The user must test with real cells to verify for themselves
how representative the model is.
As a simple guide it is useful to know:
The simulation proceeds incrementing in 0.1ml increments.
The small volume of cells and media move through kit tubing without interacting.
When cells and media enter a bag, the media is mixed with the bulk of the bag and cells are dispersed into the bag cell population.
When cells are drawn from a bag, the mixed media and cell population of the bag are supplied
When the pump is flowing in the forward direction:
The chamber is analysed as 100 segments where the cell capture/elutriate state is calculated for each cell (at that pump and centrifuge setting)
The segment capacity is calculated to estimate if the cells:
- will be held in the segment,
- overflow to the next segment or
- settle to the previous segment.
Different cell types interact in the chamber on the basis of a 'combined histogram'.
- This means larger and denser cells will occupy the chamber squeezing smaller cells further up the chamber.
Media flowing into the chamber passes through the chamber without mixing.
- The cell capacity calculation for each segment of the chamber may change as it flows through.
When the fluid flow through the chamber is reversed, the zones in the chamber containing cells and media are simply reversed out.
These interactions can be visualised once the cell simulation has completed.
Running the simulation
Once the volume simulation is complete, check the information is available to complete the cell simulation by pressing the cell Init button on the WorkFlow bar.
This will advise of any missing information needed typically bag media and cell population data.
With cell init complete press the Cell Sim arrow. The arrow will be orange to highlight it is in progress. The process can take some time � up to minutes. The duration clock illustrates progress.
Once complete final step will be highlighted Green as will the Cell Sim arrow.
The Kit View is presented in a different format and a range of visualisations are available.

This image shows the cell simulation complete. The lines in the kit, the chamber and the bubble trap display the current population.
The bag images also display the bag volume and cell population.
Black lines show where air is in the kit or bag lines.
In this case, the grey color indicates there is media in the lines and bubble trap.
The only bag with cells and a not-grey color is bag H.Clicking on Bag H reveal the contents information
It shows we have changed the media to 98% wash media, and retrieved 100% of the cells in a volume of 38.6 ml. That is a good result.
Observe the yellow kit line for valve D, this is the input line for the cells.If you click on that feature, the contents of that kit feature will be displayed. In this case a vanishingly small number of cells.
By selecting other steps in the protocol you can see where the cells have moved after each step.

Kit View Settings
The settings tab in kit view opens a window for setting visibility and colours.
Toggle active kit highlight: Changes the kit view to shadow in-active bags and lines for each step - try it.
Media only colour: Allows you to select a color contrasting between wet with media and cell populations.
Dry Fluid (Air) colour: Allow you to select a color highlighting dry segments in the kit.
For each cell type in the population:
- Hide / Show button: displays the cell type or not - allowing you to track one cell type for example.
- Colour button: Alter the base color for the cell type
Color Intensity of the display for all cells can be altered with the slider.
Animated simulation
Once the cell simulation is complete, you can view the animated movement of cells through the kit.
- You can observe the movement of the fluid through the kit.
- Air is displayed as black by default so early steps in the protocol highlight the priming actions.
- The fluid flow lines, bag contents and chamber contents are coloured according to the cell population.
- The cell groups to display and colour choices can be altered in the kit view settings.
Pick a line in the list of lines and press the play button.
You can press stop at any time during the simulation to view the status in detail. Clicking on any of the kit lines, bag lines, bags, chamber or bubble trap will display the fluid volume and contents of that feature.
Pressing Run again will re-run the same step from the start
Chamber View
Once cell simulation is complete, the chamber view tool is
enabled
 in the left-hand toolbar.
in the left-hand toolbar.

- Pick a line in the list of lines where cells are loaded or washed in the chamber.
- The chamber state displayed is the state at the end of the step.
- Press the run button to observe the changes of cell population in the chamber through the step.
- You can press stop at any time during the step simulation (there is no pause)
Chamber sub-segment population
Select a region inside the displayed chamber; the cell population is displayed.
- In this example cells are being loaded into the chamber.
- There is a concentration of T-cells and B-cells in the segment.
- These cells are washing through to a point in the chamber where they might be retained/accumulated,or they may be elutriated out if the media and chamber speed and flow rate does not support their retention.
- Granulocytes have accumulated in this subsegment but the incoming stream continues to supply Granulocytes, so any cells beyond the capacity for the segment will be elutriated through to the next segment.
- The NK cells and RBC's have 'No cap calcd' indicating the current chamber speed and flow rate will not support their retention.
Chamber Capacity plot
Press the 'Chamber Capacity' tab to plot the potential chamber retention for each cell type.

- The capacity plot highlights the location in the chamber where cells can begin to accumulate based on the centrifuge speed, pump speed and media.
- Note how some cells cannot accumulate until well into the
chamber under these conditions: media, centrifuge and pump speed.
- It does interestingly reflect observations from early bed building process development.
- Be aware these capacity estimates relate to a single cell species. That is the quoted capacity assumes there are no other cells in the system.
Total chamber contents
- Click on the chamber image in Kit view.
- The total cell population in the chamber is displayed.
- In this example a larger aliquot of the Leukopak is being been drawn in.
- The Tcell population is quoted at 100.4% reflecting the overwhelming of the chamber capacity for this cell type.
- Tcells are being lost to waste during the loading step.
- Click on Bag A to view the cells lost to Waste so far.
The simulation emulates speed ramping so intermediate points in the step can display different chamber capacity.
Guidelines
The purpose of this application is to compare a protocol, and the way it works, to a set of 'best practise' recommendations.
It is common to find guideline issues in protocols.
 This tool is enabled when the volume
simulation has been initiated.
This tool is enabled when the volume
simulation has been initiated.
If the volume simulation is completed with no guideline
conflicts, our rather concerned face becomes:
The purpose of this application is to help you the user achieve this happy face for your protocol.
Guidelines have been developed through an FMECA process which is a methodology to identify process hazards that may occur, (or have been seen to occur.)
Each hazard is documented by:
- A description of the process condition
2.The reason or hazard associated with the condition
3.How to recognise that condition in the simulation
4.Mitigation methods that might be applied
5.Where appropriate settings and/or thresholds relevant to that guideline
6.A reference number
7.FMECA Severity, Probability and Detection default values.
FMECA reporting has not yet been implemented. It will deliver a qualitative measure of protocol design improvements.
The guidelines are documented in a protected file, RoteaProtocolGuidelines_default.json
If you have loaded a protocol, the Edit Guidelines command will be illuminated. You can browse the guidelines and edit comments and settings.
If you make any changes, you will be prompted to save those changes. The default destination for the saved file is the working directory for the protocol.
The edited file 'RoteaProtocolGuidelines_Local.json', can be
applied to other protocols by copying the file to the working
directory of each protocol.
When a protocol is opened, the application checks if a local version
of the guidelines file has been saved in its working directory and
uses it. You can disable individual guideline reporting for your
applications by this method. You can also edit comments in the
guideline file and save them. This allows you to distribute your
opinions and suggested settings for the guidelines to others. (There
is no user identity or protocol content in this file.)
How do guidelines work?
- The guidelines are documented in the Rotea Protocol Guidelines file.
- Failure mode events are recognised as hard coded reviews within the process simulation.
- The purpose of the simulation is to create an environment where the guideline issues can be detected.
Example 1. Guideline AN6 Last step must be a pause step to avoid pressure fault.AlarmLevel:Error
- Review the guidelines editor for an explanation of the fault.
- This is identified by scanning the protocol for the line where 'set as last step' is set and checking it is a Pause step.
Example 2. Guideline AV13 Input bag contents dry or un-primed at start of draw down. AlarmLevel: Error
- Review the guidelines editor for an explanation of the fault.
- To recognise this event, we need to simulate the fluid flows in the protocol to understand the fluid state of the kit and bags at each step.
- To conduct the fluid simulation, information additional to the
protocol definition is required:
- The bag fluid starting volume and bag capacity
- Assignment of values to any user variables in the protocol such as harvest volume. (The minimum volume is used as a default)
Example 3. Guideline AV7 Bubble trap fails to capture bubbles from Pause loop. AlarmLevel: Warning
- Review the guidelines editor for an explanation of the fault.
- To recognise this condition, the simulator needs to know the location of air and fluids in the process kit.
- The simulator recognises air in the kit and detects the pump flow rate is above the guideline threshold setting.
- The pump flow rate threshold can be adjusted in the guideline editor.
Example 4. Guideline CL1 Cells being lost from chamber because it is overloaded. AlarmLevel: Advice
- Review the guidelines editor for an explanation of the fault.
- To recognise this condition we need to model the cells capacity to accumulate in the chamber.
- If the calculated holding capacity of the chamber is less than the number of cells, then cells will be washed out.
To conduct cell simulation we need a successful fluid simulation and further information beyond the basic protocol:
- Starting media and cell population definitions for all bags with fluid in them.
- The Rotea protocol builder can support these definitions, but they are not required for the run-time protocol.
Guideline Report tool
Reviewing guideline conflicts in your protocol
- Go to Kit view
- Select the line omn the step list of interest
- Double-click the guideline item of interest in the warning display
- The guideline report will be displayed.
Some guideline issues may occur before the volume simulation. Refer to Dealing with errors so you can proceed in these situations.
Commonly the volume simulation will trigger a number of warnings, advice and error detections.
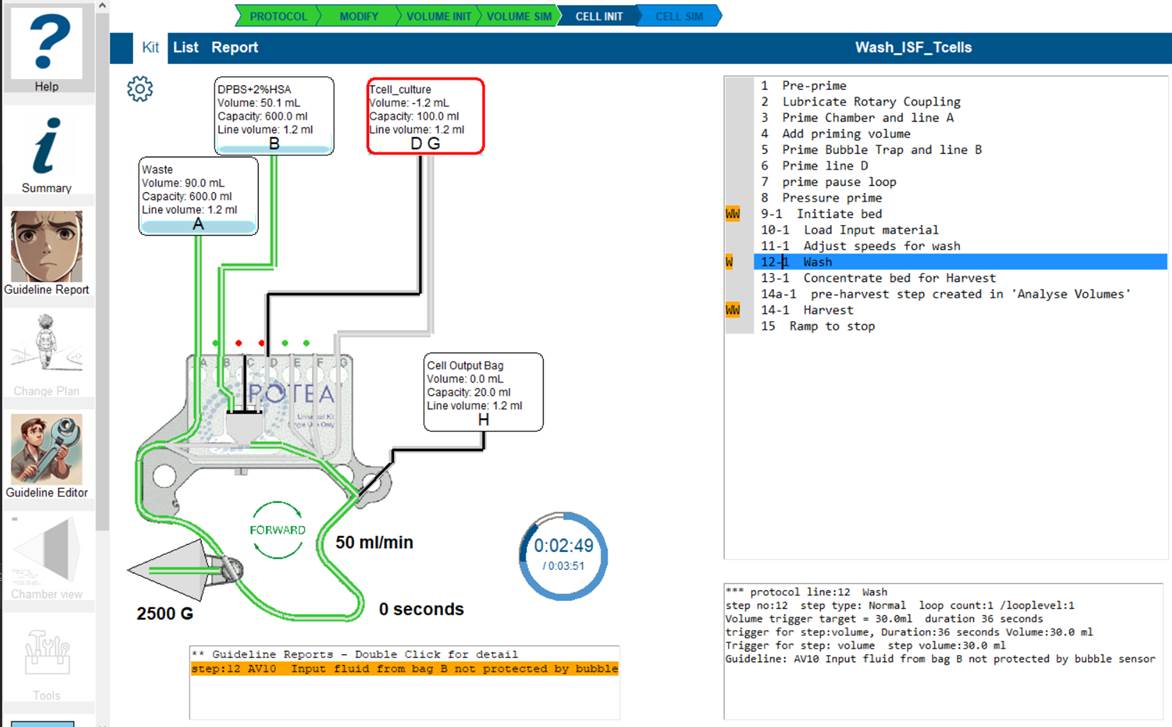
This simple protocol has successfully completed the volume simulation (Green VOLUME SIM arrow).In the line listing there are orange blocks W being displayed.
The Guideline Report tool is enabled.Click on the line of interest where an orange marker is displayed.
The warning box is displayed below the kit details with the reports for this step. Double-click on the line of interest and the Guideline report screen will open.
(You can also just click on the Guideline Report tool,this strategy opens the tool at the report you have selected.)
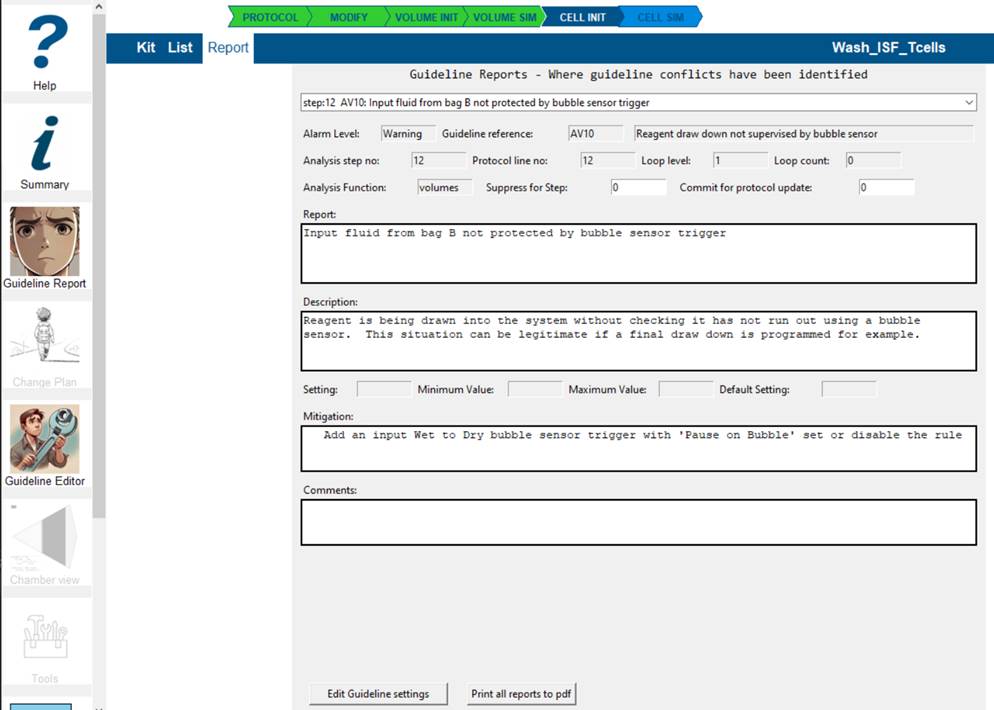
The report explains the reasoning for the conflict and allows you to:
- suppress this guideline in future for the step I am Ok with this issue remaining by setting Suppress for Step = 1 and/or
- committing this step for inclusion in changes to the protocol by setting Commit for protocol update = 1
- add comments that will be reported and transferred to any protocol change plan you propose.
You can go directly to guideline editor from here to adjust any settings or disable the guideline entirely for this protocol.
All guideline reports can be saved as a pdf by selecting that button.
The dropdown box enables you to scan the reports and select which to review.
To exit the Guideline report screen, select the Kit or List buttons to return you to those screens.Selecting Report will return to current report view.
Guideline Errors:
Guideline errors can be detected when loading a protocol, running the volume or cell simulation. The application prevents you from proceeding until you have addressed the error.
Warnings do not prevent progression of the simulations so you can observe the consequences.
Raising an error indicates this situation is not acceptable practise and likely to result in a process failure. While you cannot change the protocol or its settings, the guideline report allows you to suppress this fault for the step.
Dealing with guideline errors so you can proceed.
The guideline errors will be display in the list of lines. Go into Kit view and select the line where the error is highlighted.
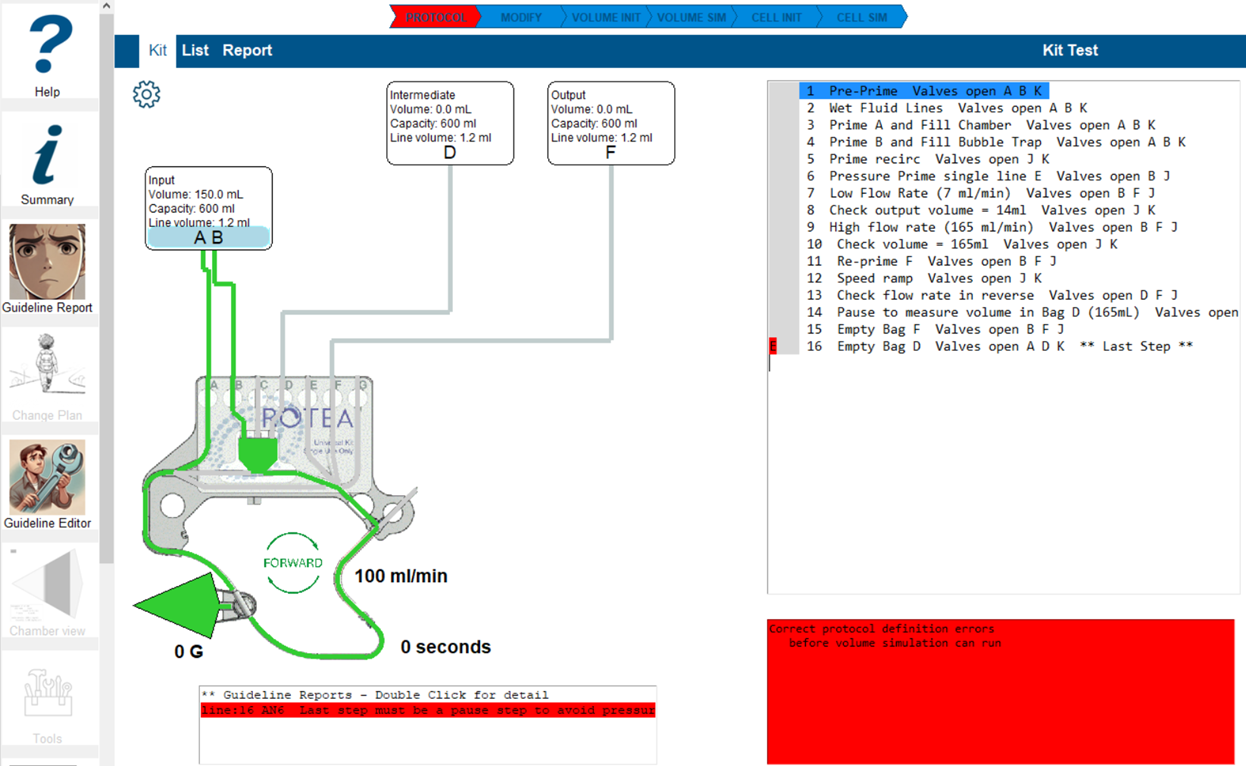
The 'Warning' box will include a highlighted line with the error summary. Double-click the line and Guideline editor will open the error report.
Change the 'Suppress for Step' entry to 1 and save the changes. This will reset the volume simulation.
Select 'Kit' view to close the Guideline editor. Select another error report, or proceed with the analysis.
The suppressed guideline report will be displayed as a warning 'S' in the simulation listing.
Guidelines you want to suppress for the entire protocol can be disabled in the 'Guideline Editor' tool
Creating a Change Plan:
 A change plan is a list of guideline reports
that have been 'committed' for change of the protocol.
A change plan is a list of guideline reports
that have been 'committed' for change of the protocol.
Delivered as a pdf file, it is designed to provide a formal decision path in situations where the protocol is a controlled document.
Adjustments to the guideline reports are retained in the Mod file but are deleted whenever a change to the protocol file is detected.Any changes to the protocol (through the Rotea protocol editor,) should include a change to its file name to preserve any change history.
The Change Plan tool is enabled when one or more guideline reports have been committed for protocol update.
All the committed guidelines can be reviewed and comments added.The change plan pdf can be generated by selecting protocol change plan to pdf.
Guideline Editor tool:
Rotea Protocol Editor:
This is freeware available from ThermoFisher Scientific at this page (not linked). Copy and paste these links in your browser to download:
DefiningMedia and Cell Populations in Rotea Protocol editor.
The protocol review application reads the protocol file to get step information including the bag data, the volumes and description or Label as described in the Bag Configuration screen of the Rotea protocol Editor.
Where possible this protocol review application tries to populate the bags with media and cell populations based on matching the Label of the bag in the Protocol Builder to standard names for media and cell populations stored in the application.
Media comprises one or more known media components mixed at a defined percentage ratio.
A cell population comprises media and one or more particles at a defined concentration per ml.
Also, if you change the media or cell data in this application, those changes are stored in the Mod file. So to introduce new changes to the RotProcRev_settings.json or from the protocol Label, then reject use of the Mod file to allow the new settings to be employed rather than the previous settings.
Rotea Software manuals:
The Best Practise protocol design principals are largely contained in these reference documents. Copy and paste these links to your browser to download:
User Guide:
Process Design Guide:
Navigation:
